Amplicon Sequencing
- Data Processing Overview
- Bulk RNA Sequencing (RNA-seq)
- Single Cell RNA Sequencing (scRNA-seq)
- Amplicon Sequencing
- Metagenomics
- Platform-Specific Microarrays
- Methylation Sequencing (Methyl-seq)
Introduction
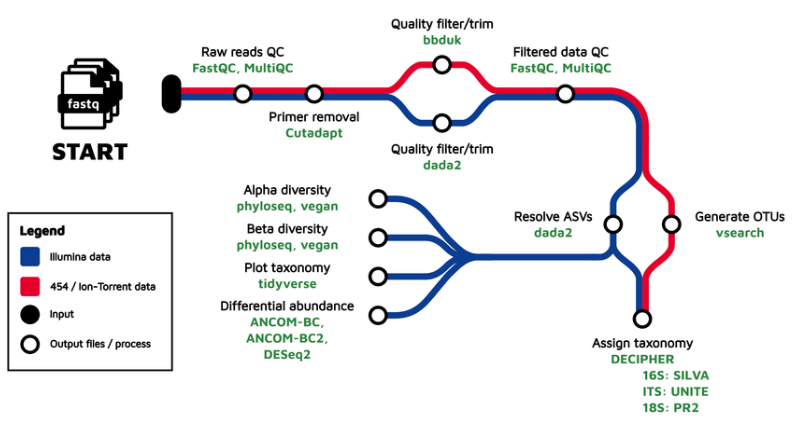
Amplicon sequencing is a targeted sequencing technique that uses specific primers to target and amplify specific genes or gene fragments from isolated DNA or RNA that has been reverse transcribed into cDNA. In microbial ecology, target genes are usually chosen due to their usefulness in delineating their source organisms in a system, and is thereby used for surveying community composition within a sample and to compare taxonomic differences across samples. Traditional analyses often cluster sequences into Operational Taxonomic Units (OTUs) based on their percent similarity, while newer approaches take into account things like sequencing error and attempt to infer the original biological sequences, often referred to as Amplicon Sequence Variants (ASVs).
The technique has broad applications across fields, from characterizing microbial diversity and analyzing environmental DNA to detecting genetic mutations in cancer research and other clinical diagnostics. Amplicon sequencing provides valuable insights into community composition, gene variation, and how these shift across treatments (such as exposure to spaceflight), environmental gradients, or time. Its cost-effectiveness and efficiency make it a powerful approach for both ecological studies and biomedical research. Within NASA’s GeneLab, the amplicon sequencing consensus processing pipeline leverages this approach to identify taxonomic changes in biological samples exposed to the space environment, highlighting its role in advancing open science and space biology.
GeneLab Data Processing Capabilities
Consensus Processing Pipeline
GeneLab worked with the scientific community via the Analysis Working Groups (AWGs) to develop a consensus pipeline for processing amplicon sequencing data hosted on the Open Science Data Repository (OSDR), to identify spaceflight-induced taxonomic changes in host and environmental samples. Each step of the pipeline and the respective output files generated are publicly available on the Amplicon Sequencing page of the GeneLab Data Processing GitHub repository.
Data Processing Workflow
The GeneLab Data Processing Team has wrapped the Amplicon Sequencing pipeline into a Nextflow workflow. This workflow is used to process all Amplicon Sequencing datasets hosted on OSDR, and the GeneLab processed data products are made publicly available alongside each dataset on OSDR. The workflow is publicly available on the GeneLab Amplicion Sequencing Workflow GitHub repository, along with instructions for how to install and run the workflow to allow users to re-process OSDR data or process their own Amplicon Sequencing data using the GeneLab workflow.