Platform Specific Microarrays
- Data Processing Overview
- Bulk RNA Sequencing (RNA-seq)
- Single Cell RNA Sequencing (scRNA-seq)
- Amplicon Sequencing
- Metagenomics
- Platform-Specific Microarrays
- Methylation Sequencing (Methyl-seq)
Introduction
Platform-specific microarray pipelines are standardized workflows developed to process and analyze data from microarray experiments, which allow researchers to study gene expression, genetic variations, or protein interactions on a chip-like platform. Microarray data are generated by isolating DNA or RNA molecules from biological samples, if RNA molecules are isolated they are subsequently reverse transcribed into complimentary DNA (cDNA), then the isolated DNA or cDNA molecules are labeled with fluorescent dyes, and hybridized to gene-specific probes fixed on a microarray slide. If the sample was expressing a known target gene, its tagged DNA or cDNA will hybridize to the respective gene probe(s) and emit a color-specific fluorescent signal upon excitation, which is detected by scanning and photographing the microarray slide. The signal intensity correlates with the degree of hybridization and therefore enables indirect measurement of gene expression. Because data quality and interpretation depend on factors such as sample preparation, array design, and scanning technology, specialized pipelines are required to ensure accurate processing of raw microarray data for each platform.
GeneLab Data Processing Capabilities
Consensus Processing Pipeline
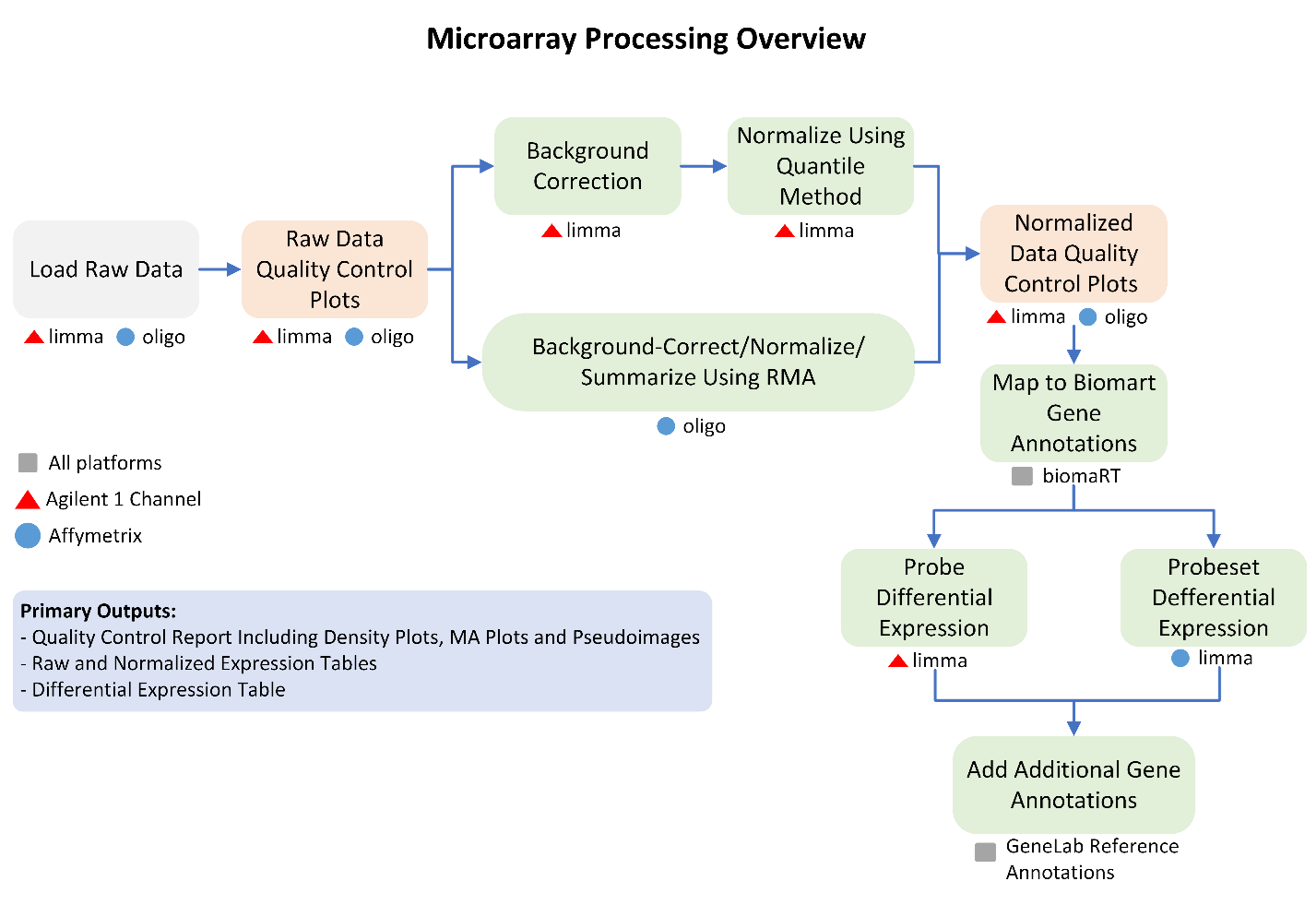
GeneLab worked with the scientific community via the Analysis Working Groups (AWGs) to develop a consensus pipeline for processing platform specific microarray data hosted on the Open Science Data Repository (OSDR), to identify spaceflight-induced differentially expressed genes. Each step of the pipeline and the respective output files generated are publicly available on the Platform Specific Microarray page of the GeneLab Data Processing GitHub repository.
Data Processing Workflow
The GeneLab Data Processing Team has wrapped the Platform Specific Microarray pipeline for the Agilent 1-channel and Affymetrix platforms into a Nextflow workflow. This workflow is used to process all Agilent 1-channel and Affymetrix microarray datasets hosted on OSDR, and the GeneLab processed data products are made publicly available alongside each dataset on OSDR. The processed data can also be visualized using the OSDR data visualization portal. The workflows are publicly available on the GeneLab Agilent 1-channel and Affymetrix Workflow GitHub repositories, along with instructions for how to install and run the workflow to allow users to re-process OSDR data or process their own Platform Specific Microarray data using the GeneLab workflow.