Methylation Sequencing (Methyl-Seq)
- Data Processing Overview
- Bulk RNA Sequencing (RNA-seq)
- Single Cell RNA Sequencing (scRNA-seq)
- Amplicon Sequencing
- Metagenomics
- Platform-Specific Microarrays
- Methylation Sequencing (Methyl-seq)
Introduction
Methylation sequencing (Methyl-Seq) is a genomic technique used to identify methylated nucleotides, a key epigenetic modification in which a methyl group is added to nucleotide residues, typically Cytosine molecules at CpG dinucleotides. These chemical modifications play an essential role in regulating gene expression, chromatin structure, and genomic imprinting. To generate Methyl-Seq data, DNA is extracted from biological samples such as cells, tissues, or whole organisms and then processed to distinguish between methylated and unmethylated cytosines before being sequenced on next-generation sequencing platforms. The resulting data provide a detailed view of the methylation state of individual bases or regions across the genome of a sample.
Applications of Methyl-Seq are wide-ranging, with particular importance in understanding gene regulation, developmental biology, disease mechanisms, and environmental effects on the genome. For example, it is used to study abnormal methylation patterns associated with cancer, neurological disorders, and autoimmune diseases, as well as to track changes in DNA methylation during development or in response to environmental exposures such as exposure to spaceflight. By offering insights into the epigenetic landscape, Methyl-Seq enables researchers to better understand how methylation influences cellular behavior, health, and disease, making it a powerful tool for both basic research and clinical applications.
GeneLab Data Processing Capabilities
Consensus Processing Pipeline
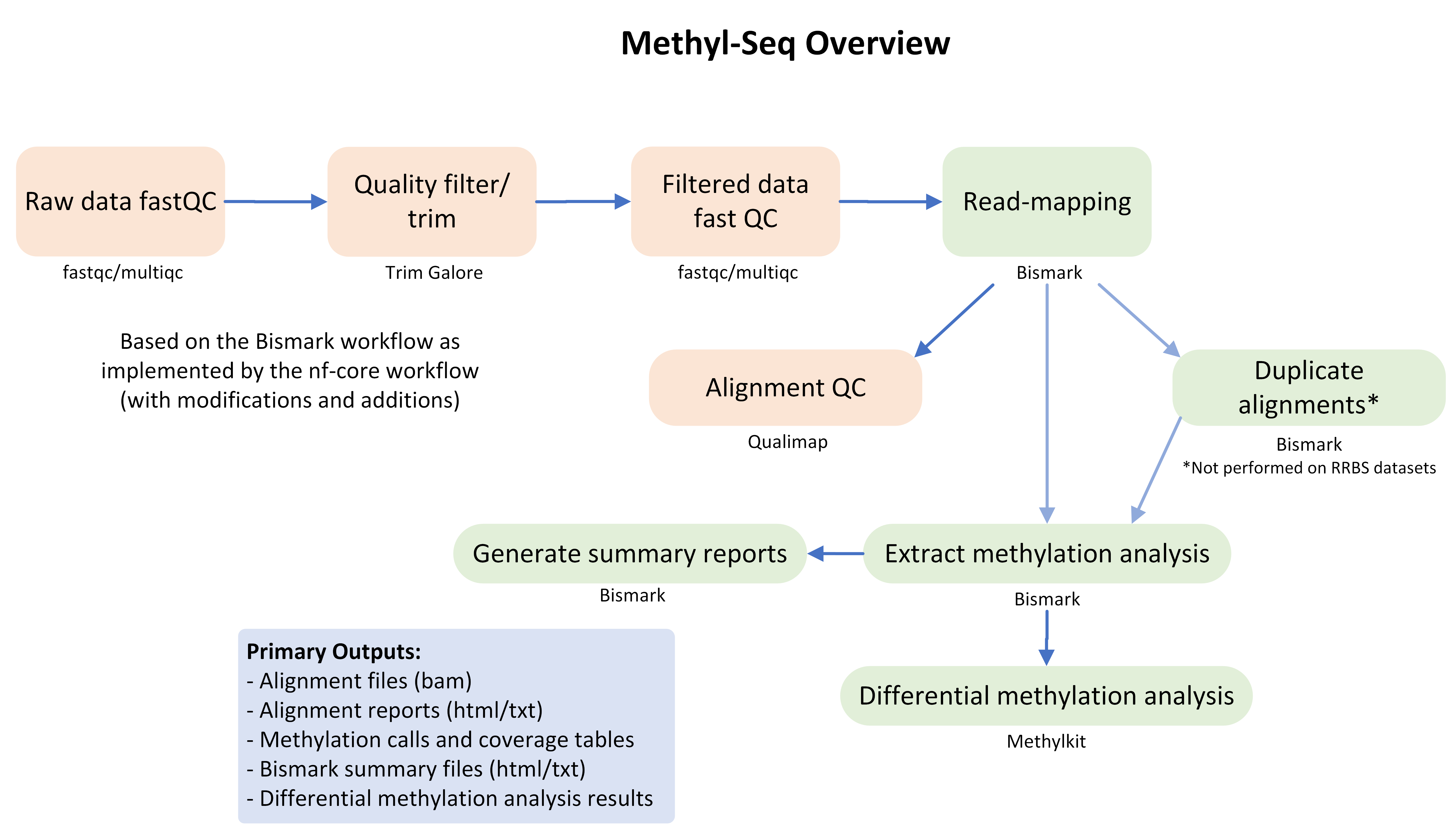
GeneLab worked with the scientific community via the Analysis Working Groups (AWGs) to develop a consensus pipeline for processing Methyl-Seq data hosted on the Open Science Data Repository (OSDR), to identify spaceflight-induced differentially methylated individual cytosine residues and regions. Each step of the pipeline and the respective output files generated are publicly available on the Methyl-Seq page of the GeneLab Data Processing GitHub repository.
Data Processing Workflow
The GeneLab Data Processing Team is currently working on wrapping the Methyl-Seq pipeline into a Nextflow workflow. This workflow will be used to process all Methyl-Seq datasets hosted on OSDR, and the GeneLab processed data products are made publicly available alongside each dataset on OSDR. The workflow will be made publicly available on GitHub, along with instructions for how to install and run the workflow to allow users to re-process OSDR data or process their own Methyl-Seq data using the GeneLab workflow.