Metagenomics
- Data Processing Overview
- Bulk RNA Sequencing (RNA-seq)
- Single Cell RNA Sequencing (scRNA-seq)
- Amplicon Sequencing
- Metagenomics
- Platform-Specific Microarrays
- Methylation Sequencing (Methyl-seq)
Introduction
Metagenomics is the study of genetic material recovered directly from host- or environmental-dervied samples, allowing researchers to analyze the collective genomes of entire microbial communities. This approach, often referred to as shotgun metagenomic sequencing, amplifies all accessible DNA within a mixed community to reveal both taxonomic diversity and functional potential. One powerful outcome of metagenomics is the recovery of Metagenome-Assembled Genomes (MAGs), which represent reconstructed genomes of closely related microbial populations, which could match an existing isolate or represent a novel isolate, including unculturable species. The ability to generate MAGs has significantly expanded the Tree of Life and provided unprecedented insights into microbial ecology and evolution.
The applications of metagenomics span numerous fields. In environmental microbiology, it reveals the diversity and roles of microbes in ecosystems, such as the space environment, and biogeochemical cycles. In human health, it deepens our understanding of the microbiome’s impact on disease, immunity, and metabolism. In biotechnology and industry, it facilitates the discovery of novel enzymes, antibiotics, and bioactive compounds, while in disease surveillance, it helps track pathogens and outbreaks. By capturing both taxonomic structure and functional potential across space, time, and treatments, metagenomics provides a transformative lens into microbial communities. Advances in sequencing and computational tools continue to expand its potential, making it an essential approach for tackling challenges in science, medicine, and industry.
GeneLab Sample Processing Standard Operating Procedures
GeneLab provides publicly available Standard Operating Procedures (SOPs) for sample extraction, library preparation, and sequencing steps using different approaches and kits. Researchers can access these protocols and use them to process samples in their own laboratories. Explore and download the GeneLab Sample Processing SOPs on GitHub.
GeneLab Data Processing Capabilities
Consensus Processing Pipeline
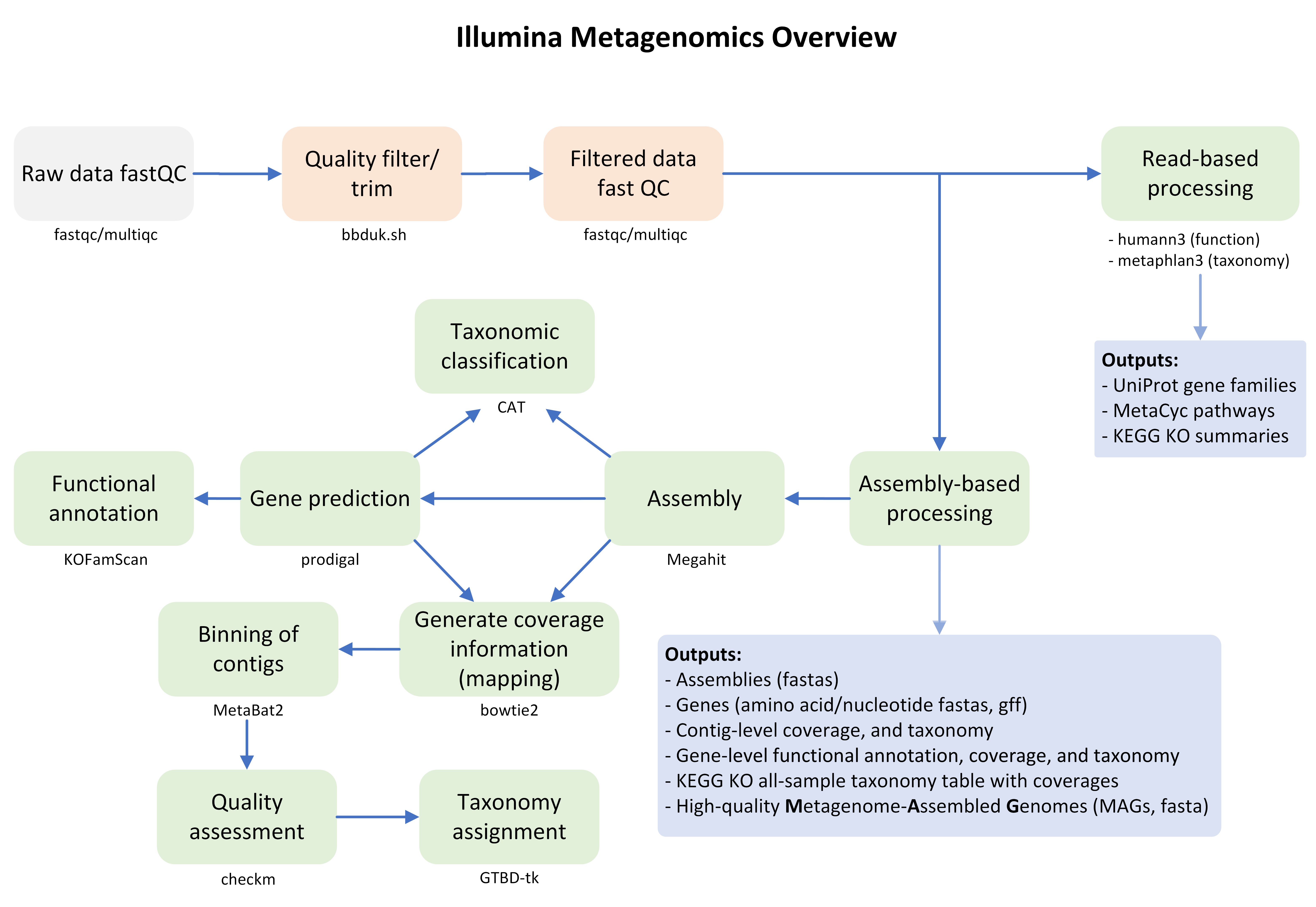
GeneLab worked with the scientific community via the Analysis Working Groups (AWGs) to develop a consensus pipeline for processing shotgun metagenomic sequencing data hosted on the Open Science Data Repository (OSDR), to identify spaceflight-induced taxonomic and functional changes in host and environmental samples. Each step of the pipeline and the respective output files generated are publicly available on the Metagenomics page of the GeneLab Data Processing GitHub repository.
Data Processing Workflow
The GeneLab Data Processing Team has wrapped the Metagenomics pipeline into a Nextflow workflow. This workflow is used to process all Metagenomics datasets hosted on OSDR, and the GeneLab processed data products are made publicly available alongside each dataset on OSDR. The workflow is publicly available on the GeneLab Metagenomics Workflow GitHub repository, along with instructions for how to install and run the workflow to allow users to re-process OSDR data or process their own Metagenomics data using the GeneLab workflow.