Bulk RNA Sequencing (RNA-seq)
- Data Processing Overview
- Bulk RNA Sequencing (RNA-seq)
- Single Cell RNA Sequencing (scRNA-seq)
- Amplicon Sequencing
- Metagenomics
- Platform-Specific Microarrays
- Methylation Sequencing (Methyl-seq)
Introduction
Bulk RNA sequencing (bulk RNA-Seq) is a widely used technique in molecular biology that measures gene expression in a sample, such as cells, tissues, or whole organisms. The process involves converting RNA molecules into complementary DNA (cDNA) and sequencing them using next-generation sequencing platforms. Because ribosomal RNA (rRNA) makes up more than 80% of total RNA and is usually not the focus of these studies, it is removed during sample preparation prior to cDNA conversion, typically through ribo-depletion or by selecting for messenger RNA (mRNA) using polyA-selection. Bulk RNA-Seq provides powerful, large-scale insights into gene expression, enabling comparisons between different conditions (e.g., healthy vs. diseased or spaceflight vs. ground control), discovery of new transcripts, biomarker identification, and pathway analyses. While it offers broad, quantitative profiling, the method averages signals across many cells, which means it cannot capture single-cell differences, and the resulting data can be complex and costly to analyze.
GeneLab Sample Processing Standard Operating Procedures
GeneLab provides publicly available Standard Operating Procedures (SOPs) for sample extraction, library preparation, and sequencing steps using different approaches and kits. Researchers can access these protocols and use them to process samples in their own laboratories. Explore and download the GeneLab Sample Processing SOPs on GitHub.
GeneLab Data Processing Capabilities
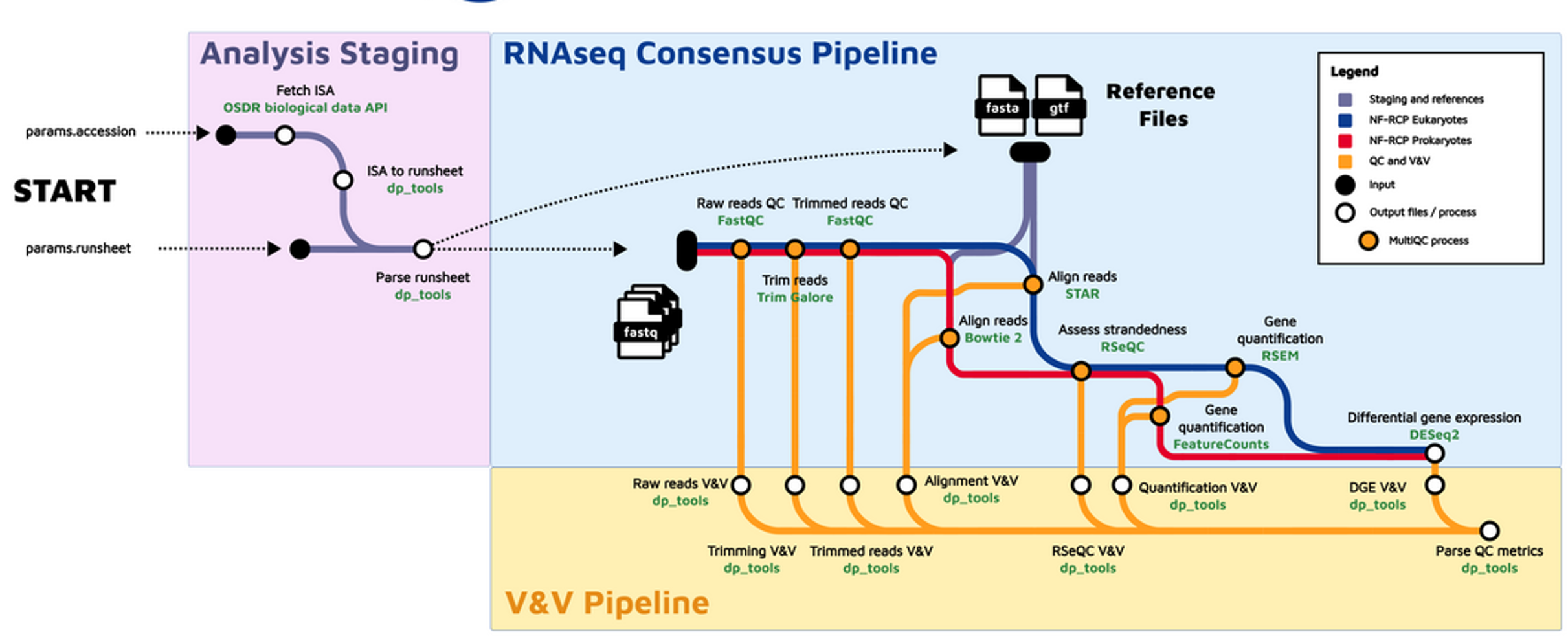
Consensus Processing Pipeline
GeneLab worked with the scientific community via the Analysis Working Groups (AWGs) to develop a consensus pipeline for processing bulk RNA-Seq data hosted on the Open Science Data Repository (OSDR), to identify spaceflight-induced differentially expressed genes. Each step of the pipeline and the respective output files generated are publicly available on the bulk RNA-Seq page of the GeneLab Data Processing GitHub repository.
Data Processing Workflow
The GeneLab Data Processing Team has wrapped the bulk RNA-Seq pipeline for eukaryotic and prokaryotic organisms into a Nextflow workflow. This workflow is used to process all bulk RNA-Seq datasets hosted on OSDR, and the GeneLab processed data products are made publicly available alongside each dataset on OSDR. The processed data can also be visualized using the OSDR data visualization portal. The workflow is publicly available on the GeneLab RNA-Seq Workflow GitHub repository, along with instructions for how to install and run the workflow to allow users to re-process OSDR data or process their own bulk RNA-Seq data using the GeneLab workflow.